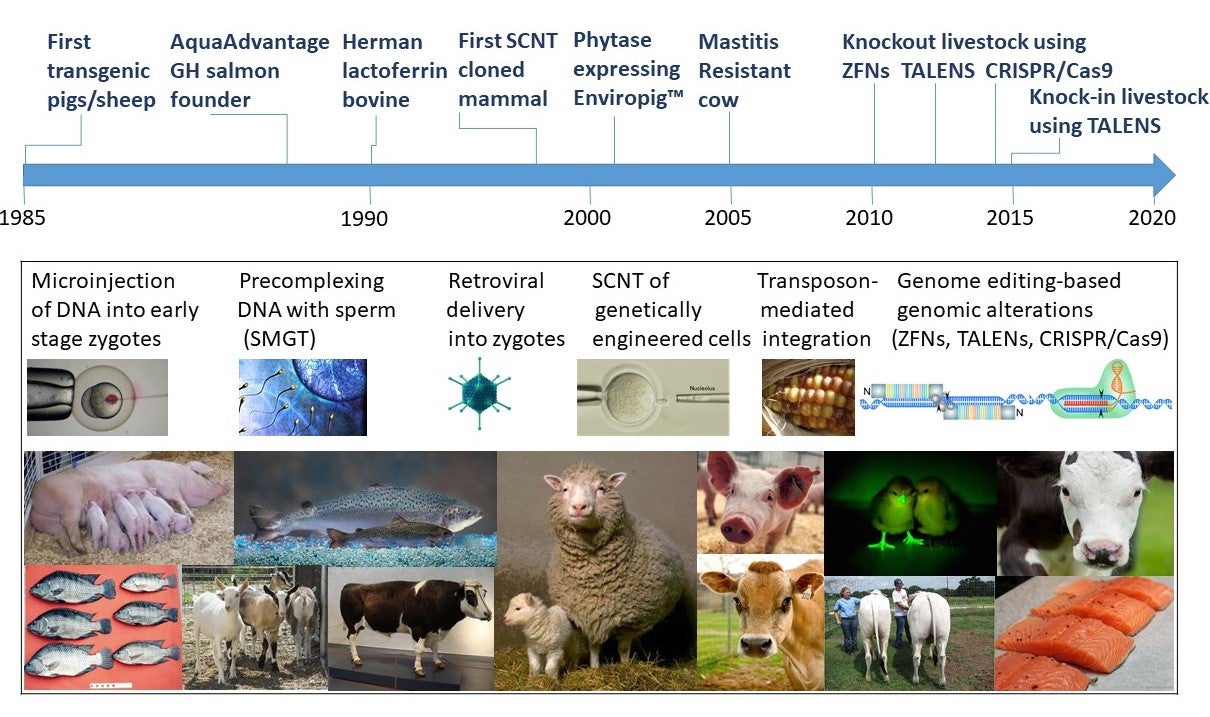
This is part 3 of a 1, 2, 3 part series on Genetic Engineering in Livestock
In 1996, the year of Dolly the infamous first somatic cell nuclear transfer (SCNT) cloned mammal and the promulgation of the United States Coordinated Framework for Regulation of Biotechnology , a review paper found that of the 289 published papers in PubMed when searching for the term “transgenic livestock”, the vast majority of original reports focused on growth enhancement, and that 24% of them were review articles. This proportion has not changed much, with that same search today returning approximately 30% review articles. What happened to the promise of transgenic livestock that almost one quarter of the publications in the field are reviews rather than original research articles, and only a single transgenic food animal has been commercialized in 35 years (as discussed in Part 2 of this series)?
The livestock breeding sector is distinct from the plant breeding sector where transgenic plants are grown by over 17 million farmers globally. Some of this is due to biological differences between the two kingdoms, including the mode of reproduction (e.g. plants can self-pollinate), and the number of progeny per reproduction cycle. A major difference between animal and plant breeding is that the former places greater emphasis on population improvement, with product development consisting primarily of multiplication of these improved genetics through outcrossing, whereas in the latter greater emphasis is placed on selection of an improved product in the form of a recognizable plant variety, which is often also the source of parents for the next breeding cycle .
One transgenic plant transformation event (often protected by IP) can be amplified in seed multiplication, but in animals the founder event will have to occur in elite stock in the breeding nucleus and then be transmitted down to the commercial sector through multipliers, a process that can take decades depending upon the generation interval of the species.
Additionally, desired traits will need to be introduced into several breeds or lines in animal breeding programs because they often make extensive use of heterosis (or hybrid vigor) through crossbreeding. For example, a typical modern broiler chicken breeding program is shown in Figure 1.
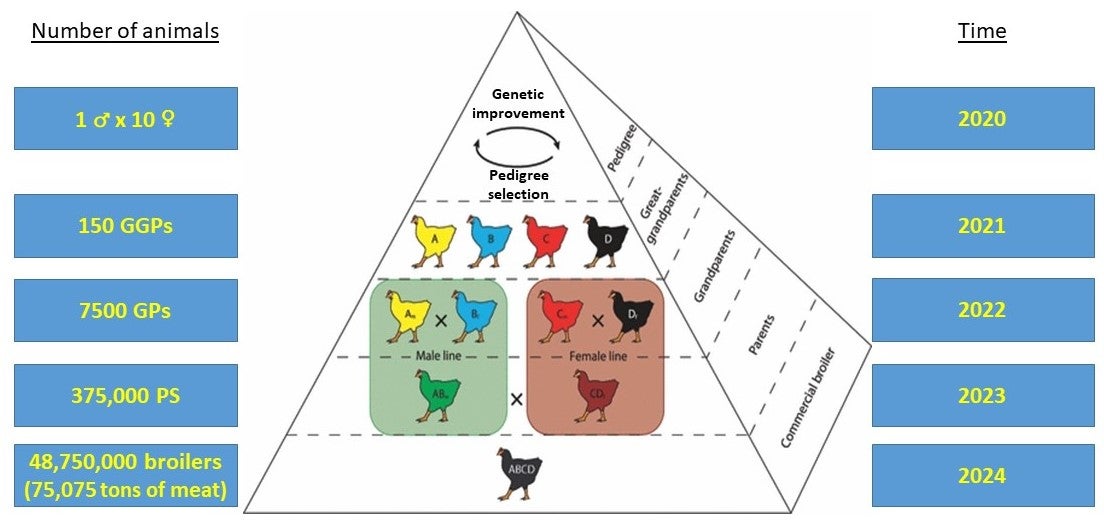
Figure 1. A typical modern broiler chicken breeding program, represented as a pyramid where each level represents a generation, and the approximate timeline to move genetics from top of the broiler breeding pyramid to the consumer. (Modified)
Genetic improvement and pedigree selection for desired traits occurs at the top of the pyramid across four lines. Within the pedigree segment there are specific male and female lines, with the males typically selected for heritable growth and production traits and the female lines selected for early growth and conformation. The commercial broiler (5th generation) at the bottom of the pyramid is a four-way cross derived from the cross of a male and female parent line.
Genome editing offers an efficient approach to introduce useful genetic variation into livestock breeding programs through targeted inactivation of gene function, and/or through allele introgression in the absence of undesired linkage drag. Genome editing involves the use site-directed nucleases (e.g. TALENS (Transcription Activator-Like Effector Nucleases) or the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/CRISPR-Associated Systems (CAS)) to efficiently introduce a double stranded break (DSB) at a predetermined location in the genome. The DSB can be repaired by the cell’s natural DNA repair mechanism (non-homologous end joining; (NHEJ)), often resulting in single nucleotide changes, deletions or small (1-2 nucleotide) insertions at the DNA cut site. In this case, although the location of the cut site is very precise, the exact change that occurs when the DNA is repaired is random and so a number of different outcomes representing minor sequence changes are possible.
Alternatively, repairs can be directed to introduce, delete, or replace a series of letters in the genetic code using a nucleic acid template. This essentially enables the introduction of known, desired alleles or haplotypes via homology directed repair (HDR) based on what is understood about naturally-occurring genetic variation in the target species. It is now possible to alter animal genomes without necessarily incorporating transgenic genetic material. To date SCNT cloning of edited somatic cells, especially HDR donor-template defined alterations, has been the primary method to produce livestock carrying nuclease-mediated genetic changes, especially HDR donor-template defined alterations, in their genomes.
Researchers have already produced a number of gene-edited farm animals for biomedical research applications, and also for agricultural purposes (see Table 2 in my recent review paper). The latter group includes animals carrying no novel DNA sequences (e.g. Porcine Reproductive and Respiratory Syndrome (PRRS) virus-resistant CD 163 knockout pigs ; myostatin knockout sheep and cattle with increased lean muscle yield ; and knockout sheep with increased wool length and yield ; intraspecies allele substitutions (e.g. hornless dairy cows due to an allele substitution at the POLLED locus ); intraspecies gene insertions also known as cisgenics (e.g. cows with increased resistance to tuberculosis due to knock-in of bovine NRAMP1 gene); and animals with interspecies allele substitutions, analagous to traditional transgenic applications (e.g. domestic pigs (Sus scrofa) carrying an allele from the African Swine Fever-resistant African warthog (Phacochoerus africanus).
Not surprisingly, the traits that have been targeted by researchers with gene editing (animal health and well-being, product quality and yield) are common to the breeding objectives of traditional selection programs. Health and welfare traits are also of particular interest to the general public, especially projects like the female-only layer chickens. As with earlier genetic engineering approaches, whether breeders will be able to employ gene-editing in commercial farm animal genetic improvement programs will very much depend upon global decisions around regulatory frameworks and governance.
A chance to rethink
In early 2017, the United States Food and Drug Administration (FDA) released its updated draft “Guidance for Industry #187” and changed the title to “Regulation of Intentionally Altered Genomic DNA in Animals”. This guidance proposes to regulate all food animals whose genomes have been intentionally altered using modern molecular technologies, including gene editing technologies, as new animal drugs. This includes many of the same nucleotide insertions, substitutions, or deletions that could be obtained using conventional breeding. No longer is it the presence of a transgenic rDNA construct that triggers mandatory premarket FDA regulatory oversight prior to commercial release, but rather it is the presence of any “intentionally altered genomic DNA” in an animal’s genome that initiates oversight.
This runs against the wording of the 1986 Coordinated Framework which indicated regulatory review was only required for organisms deliberately formed to contain an intergeneric (i.e. transgenic) combination of genetic material. It also runs counter to the OSTP policy, the 2016 U.S. National Academies of Sciences, Engineering, Medicine report, which recommended a “product not process” regulatory trigger. In addition it is contrary to the USDA approach to the regulation of gene edited food plants, and is also out of step with decisions being made by other regulatory agencies in a number of countries around the world (e.g. Argentina), with implications on global trade. Mandating premarket regulatory approval for deletions, mutations, and the conversion of one wild-type allele to another wild-type allele in the same species (cisgenic) that could have been obtained using conventional breeding is not scientifically defensible, given the known genetic variation that exists naturally in livestock genomes.
Figure 2 shows the global regulatory landscape for gene-edited animals. If those animals do not carry novel DNA (transgenic DNA), meaning that the DNA alteration could have been achieved using conventional breeding, then a number of countries are not requiring additional regulatory oversight. To date only a single entity, a large global animal genetics company, has announced plans to commercialize a gene edited food animal. The University of Missouri signed an exclusive global licensing deal for potential future commercialization of PRRS virus-resistant pigs with UK-based Genus plc. This company has also entered into a strategic collaboration with Beijing Capital Agribusiness Co. Ltd, a leading Chinese animal protein genetics business, to research, develop, register and market elite pigs that are resistant to PRRS virus in China.
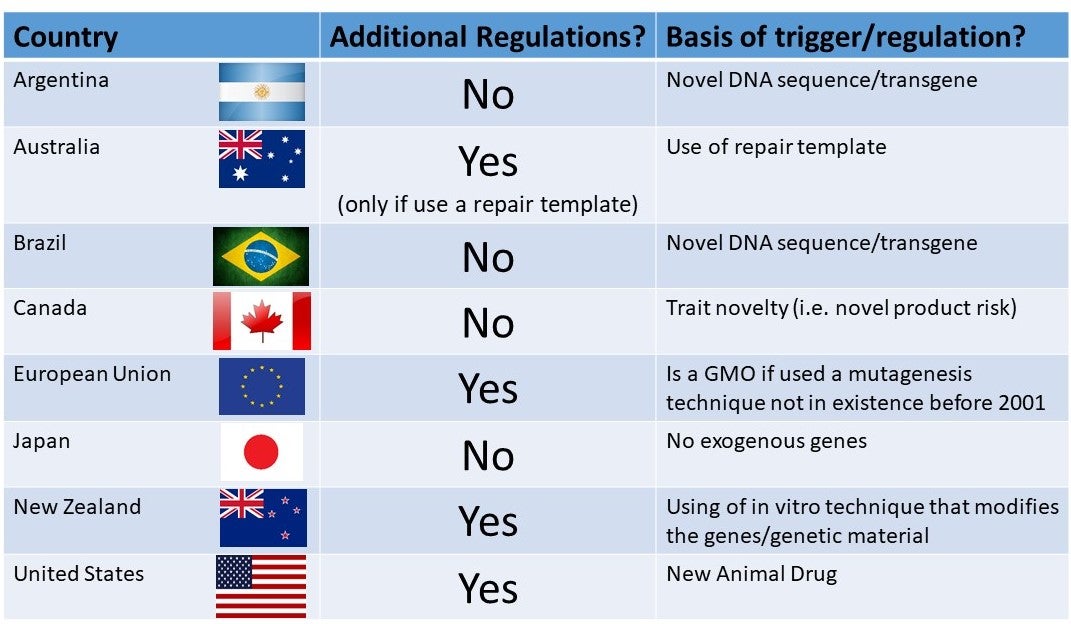
Figure 2. Global picture of whether additional regulations will be triggered for gene-edited animals that do not carry transgenic DNA
In a February 2020 correspondence entitled Genome editing in animals: why FDA regulations matters published in Nature Biotechnology, FDA Center for Veterinary Medicine (CVM) Director Steven M. Solomon made the case for “why it is necessary for there to be regulatory oversight of intentional genomic alterations in animals, even when the intended modification seeks to replicate a naturally occurring mutation.” He then specifically distances his argument from intentional genomic alterations performed in organisms from other kingdoms that we eat, i.e. plants and microbes, with the statement “Readers should note that our statement here relates to intentional genomic alterations in animals; we are not commenting on alterations in plants or other organisms.”
This suggests there is something uniquely risky about intentional genomic alterations in animals. However, the history of plant breeding is rife with examples where antinutrients in plants have inadvertently been increased through conventional selective breeding efforts (e.g. think solanine in potatoes, or psoralens in celery). I have a hard time coming up with an analogous food safety concern from conventional animal breeding. Dr. Solomon then goes on to support his argument with the a case of a naturally-occurring genomic variant that was deleterious to animal health and that increased in frequency due to conventional selection. “There is a particularly compelling example of the risks of occult genomic alterations in cattle produced by traditional breeding techniques: a high incidence of bovine leukocyte adhesion deficiency (BLAD) syndrome, a lethal autosomal recessive disease, in Holstein calves.”
In an unusual move, Nature Biotechnology published its own editorial response the FDA’s correspondence countering that, “If the BLAD case history tells us anything, it is that the origin of a DNA arrangement (conventional breeding, recombinant DNA or gene editing) makes little difference to an animal. The genomes of domestic cattle contain millions of natural variants: the 1000 Bull Genomes Project found >86.5 million differences (insertions, deletions and single nucleotide variants) among cattle breeds. According to prominent researchers in the field, none of these variants has been shown to produce ill effects on consumers of milk or meat. Amidst this background of innocuous variation, how can the presence of one identifiable variant justify the costs and delays of mandatory FDA oversight?
“The US Food and Drug Administration is sticking to its plan to carry out mandatory premarket review of all gene-edited livestock, irrespective of trait risk. It should rethink.
The US Food and Drug Administration (FDA) Center for Veterinary Medicine (CVM) continues to argue that every animal created by gene editing should be subject to mandatory premarket review and substantial safety testing. This position, first outlined in a 2017 draft guidance, presupposes that all gene-edited animal are potentially hazardous — despite the evidence and the views of many researchers. It canonizes a precautionary stance to gene-edited animals, irrespective of the trait engineered, even if the animal is indistinguishable from one created by conventional breeding. It is out of step with the practice of the US Department of Agriculture (USDA), which recently deferred from regulating gene-edited products. And it goes against decades of US policy focusing on appropriate product risks and benefits through the Coordinated Framework.”
Course Correction editorial, Nature Biotechnology, February 2020
I have spent a career in animal genetics and genomics. For a good chunk of that time, it has not been possible to use the tools of molecular biology to introduce useful genetic variation like disease resistance into livestock breeding programs through transgenesis. Genome editing offered a new opportunity to use molecular tools in a way that would not necessarily introduce transgenic DNA. It was hoped that this would help avoid the regulatory costs that effectively strangled transgenic applications like the USDA ARS’ mastitis-resistant cow, and University of Guelph’s “Enviropig“. Precluding these transgenic animals from coming to market is associated with a forgone opportunity cost as we have not cured mastitis, and pigs still can’t digest phytate. Genetic solutions that could have helped to address these animal disease and phosphorus pollution problems have just been sidelined.
There was some hope that gene-edited animal alterations that could have been achieved using conventional breeding, would not be treated differently to conventional breeding from a regulatory standpoint. This was the decision of the USDA regarding genome edited plants, and that of several countries (Figure 2). However, the decision in some countries including the US, is to regulate gene-edited animals based on the fact they are gene-edited, rather than product novelty or risk. I fear this will lead to another long list of forgone genetic innovations in US animal agriculture, and advantage countries with a product-risk focused regulatory approach.