In what seems like a scene from the movie Groundhog Day, another rat study has come out of the laboratory of Dr. Giles-Eric Séralini, only in this case it is Roundup and not GMOs that are under fire. When I read the title of the paper, “Multiomics reveal non-alcoholic fatty liver disease in rats following chronic exposure to an ultra-low dose of Roundup herbicide”, I assumed a new study had been performed by the laboratory showing what this specific title appears to conclude i.e. that rats exposted to low levels of Roundup developed non-alcoholic fatty liver disease. However, when I read further I found that this was a study on tissues from a subset of the same lumpy rats that were involved in the famously retracted (and subsequently republished) paper from 2012 – the rats with horrific tumors (not fatty livers) due to GMOs (not glyphosate) that was breathlessly reported on the Doctor Oz show I participated in, and by media throughout the world.
I think if my work had been roundly criticized by scientific peers for poor experimental design and pathology data inadequacies, and critiqued by a multitude of separate national biosafety committees from Belgium, Brazil, European Union, Canada, France, Germany, Australia/New Zealand, and The High Council on Biotechnology, I would not double down and continue to analyze 5-year old samples from that same experiment. What is weird is that although I vividly remember the images of grotesque tumors on the white Sprague Dawley female rats, (one does not forget those images with a “GMO” label contrasted against the shocking tumors) I did not recall any mention of non-alcoholic fatty liver disease. So I went back to the original paper and searched for the term “fatty liver disease”. Nada.
In fact, the only data on livers in that retracted/republished 2012 paper was presented for the male rats. According to the 2012 paper the males that received the low levels of Roundup (50 ng/L glyphosate equivalent dilution) displayed liver “congestions” and “macroscopic and microscopic necrotic foci”, not fatty liver disease. I asked a Laboratory Animal pathologist at UC Davis who specializes in rodent health to review the data in the paper to determine if it suggested the rats had fatty liver disease. There was no histopathologic evidence of hepatic lipidosis presented in males and no data on female livers was presented at all. Many of the “anatomical pathologies” observed are common aging related findings and this was not taken into account or discussed. They suggested the term “anatomopathological analysis” was a very irregular term for a veterinary pathologist to use and that the use of hepatodigestive tract and liver as separate categories of pathology incidence were redundant. They kept doggedly going back to the fact that no fatty liver phenotype data were ever presented on female livers so they could make no determination as to whether or which rats were suffering from fatty liver disease.
If you want a really interesting read from a group of veterinary pathologists who reviewed the pathology data in the 2012 Séralini study, their review contains the following understated scientific barbs (bold emphasis mine).
“The sentence ‘The largest palpable growths (…) were found to be in 95% of cases non-regressive tumors, and were not infectious nodules.’ is very confusing. We hope that differentiating inflammatory from neoplastic lesions was not a challenge for the authors. Another clear example illustrating the lack of accuracy of the results is found in Fig. 3 where microscopic necrotic foci in the liver are grouped with clear-cell focus and basophilic focus with atypia. The first finding refers to a degenerative process whereas the remaining two refer to a proliferative one (Thoolen et al., 2010). Such basic error would be considered as a disqualifying mistake at an examination for pathologists.”
Ouch.
They then go on to ask why there was no mention of which pathologist did the analyses, and why the rats were not euthanized earlier
“as most members of the ESTP [European Society of Toxicologic Pathology] are veterinarians, we were shocked by the photographs of whole body animals bearing very large tumors. When looking at the lesions, we believe those animals should have been euthanized much earlier as imposed by the European legislation on laboratory animal protection”
and then conclude their diatribe with the following
“The ESTP comes to the conclusion that the pathology data presented in this paper are questionable and not correctly interpreted and displayed because they don’t concur with the established protocols for interpreting rodent carcinogenicity studies and their relevance for human risk assessment. The pathology description and conclusion of this study are unprofessional. There are misinterpretations of tumors and related biological processes, misuse of diagnostic terminology; pictures are not informative and presented changes do not correspond to the narrative.”
For those who are not immersed in science – these are damning criticisms.
So back to the 2017 study which cites a 2015 “transcriptomics” study by the same group for the observations on the female livers. In that study livers from 10 control females and the 10 females from the R (A) group from the 2012 study (for those of you paying attention) were analyzed using “transcriptomics”. So I went to read the 2015 paper to see if the Roundup-ingesting females perhaps had some liver data, and again there was no discussion of a fatty liver disease phenotype. There was however, an interesting discussion of why tissues from the females was used for the analysis in both the 2015 “transcriptomics” and 2017 “multiomics” paper.
In the 2012 study that started it all, apparently
“Most male rats were discovered after death had occurred. This resulted in organ necrosis making them unsuitable for further analysis. We therefore focused our investigation on female animals where freshly dissected tissues from cohorts of 9-10 euthanized and untreated rats were available. Female control and Roundup-treated animals were respectively euthanized at 701 ± 62 and 635 ± 131 days. Anatomopathological analysis of organs from these animals revealed that the liver and kidneys were the most affected organs.”
Well, the fact that the males got to a stage of necrosis because no one discovered they were dead seems strange in a study where rats are presumably checked every day as required by every animal care protocol I am familiar with. However, such protocols would also have required the rats to be sacrificed long before the tumors were able to grow to the size that were clearly evident in the photos associated with this study. And the fact that the liver and kidneys were the most affected organs might well have been true for the male rats (and these apparently necrotic tissues were analyzed and reported for these males), but for the female rats, according to the 2012 paper, it was all about the tumors!
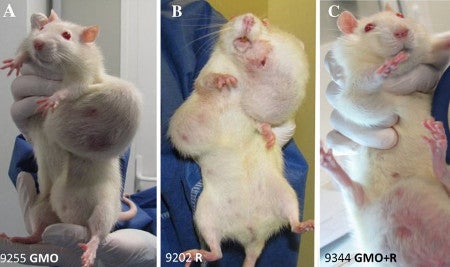
Image from Seralini et al. 2012
That was the whole basis of the sensational 2012 paper that actually resulted in entire African countries rejecting all GMO imports. Reread that previous sentence because it shows the power of this one, poorly-designed study with 120 rats.
So livers were being harvested from these 20 females – several of which were compromised and euthanized “early” (2 from the control group, and 5 from the “treatment” group) at different ages due to the tumor load. Is it not obvious that these additional factors of tumor load and different ages would confound any data collected from their livers?
The 2015 paper goes on to show electronic microscope analysis of liver sections from females. But it turns out the photograph of the control female was actually the same photo at a different magnification as that shown for the control male hepatocyte image in the 2012 paper. The authors have since stated that was an honest mistake and have submitted a corrigendum, but go on to suggest that there are differences in the hepatocytes from Roundup-treated rats, specifically showing “a disruption of glycogen dispersion”, a disruption of nucleolar function and an overall decreased level of transcription. How transcription can be determined based on an electron micrograph is unclear. No mention is made of the “fatty liver disease” promised in the 2017 paper’s title.
So let me sum this up for those of you who may be lost. The original, highly-controversial 2012 study was done on 120 rats. The most recent study was performed on the livers of a subset of 10 female control rats and 10 female rats from that same 2012 study that were in “Roundup group (A)” which received 50 ng/L glyphosate equivalent dilution in their water. We do not know their water intake so have no idea of actual dosage of “Roundup”; we have little histological data on female liver samples – let alone a diagnosis of fatty liver disease; we know that the control and “treated” rats were euthanized at a variety of differing ages, and that the majority of these female rats had huge tumors that required several of the rats in both the control and Roundup groups to be euthanized before two years of age. And the livers from these 20 rats were the basis of the most recent “omics” paper. There is a saying in science (and perhaps other disciplines): “garbage in – garbage out”.
So let’s plough on – and read the 2017 paper which concludes that the metabolome and proteome analyses of the livers from the “Roundup-drinking” rats versus the controls “showed a substantial overlap with biomarkers of non-alcoholic fatty liver disease and its progression to steatohepatosis”. Hooray – now THERE is a testable hypothesis – so what ARE the biomarkers of non-alcoholic fatty liver disease? In other words, what proteins and metabolites might you expect to see upregulated (or downregulated) if in fact animals had non-alcoholic fatty liver disease? I have read the paper several times now and seen no reference to a paper that answers that question. So in the absence of knowledge of fatty liver biomarkers, and given the fact no pathology diagnosed “fatty liver disease”, to conclude that “Multiomics reveal non-alcoholic fatty liver disease in rats following chronic exposure to an ultra-low dose of Roundup herbicide” is – to put it kindly – overstating the results of the research and making conclusions beyond that supported by the data.
Interestingly the bioinformatics analysis in this 2017 paper appears to be an improvement on previous works by this group in that the p-values were adjusted to account for the fact there were a high number of metabolites measured (1906 proteins and 673 metabolites), and there was therefore a need to do corrections for multiple comparisons to try to minimize the number of false positives. The authors even include a discussion of the need for corrections for multiple comparisons on page 9, and correctly state that there is a need to do this when measuring hundreds or thousands of observations to reduce the chance of making a type I error (false positive). However, they then lament the fact that there was a lack of statistical significance following the multiple comparison correction for all but three metabolites, due to the small sample size. That is the point! That is why these studies need to have sample size determinations based on the hypothesis being tested.
This study, which was based on the experimental design of a 90 day subchronic toxicity study (OECD, 1998) such that 10 animals were assigned to each group, was critiqued by the German Federal Institute for Risk Assessment (BfR) for small sample size for that very reason.
“subchronic studies show a substantially lower variation of age-related pathological changes between animals within a group while those changes are inevitable in long-term studies. As the published study has confirmed, the two-year duration of the study is of the order of the expected life span in rats including the Sprague Dawley strain that was used in the study. This strain, provided by the breeder Harlan, is known to develop spontaneous tumors, particularly mammary and pituitary tumors, at relatively high rates compared to other strains (Brix et al., 2005; Dinse et al., 2010). Therefore, it can be expected that a significant number of animals develop age-related illnesses or die for diverse reasons already during conduct of the study. The distribution of the cases of death between groups can be random, and a number of 10 animals per sex and group is too low to confirm a trend or an effect. Furthermore, no statements on statistically significant dose-response-relationships can be made. Larger sample sizes, as recommended for carcinogenicity studies in OECD Test Guidelines No. 451 or No. 453, would be required in order to allow precise statements with respect to the findings.”
In other words you need to have bigger sample sizes to perform long term studies because many changes are associated with old age – especially when working with a rat strain that is known to develop spontaneous tumors, particularly female mammary and pituitary tumors!
Frustratingly, when the multiple comparisons removed all but three of the 673 metabolites as being statistically significant due to multiple comparison correction in the 2017 paper, the authors just went ahead and included the 55 that had a significant uncorrected p value(!), because “the non-adjusted statistically significant levels” fit a narrative, and so were revived from the statistical trash can on the basis that “they were found to be non-random and thus biologically meaningful”. This is the very definition of confirmation bias which is what multiple comparison correction and correct experimental design is trying to weed out because scientists are people too, and they are not without their own preconceived notions of how the world works.
More concerning, this 2017 paper is yet another in a string of papers from this group that was accepted in a peer-reviewed journal, in this case Scientific Reports, an online journal from the publishers of Nature. The problems in experimental design, lack of supporting pathology data on the test subjects, and wildly subjective overinterpretation of the results should have been grounds for soundly rejecting this manuscript. We live in an age of the willful neglect of scientific evidence, and the emergence of “alternative facts” and realities. As a scientist it worries me that papers like this are published in apparently respected journals. I remember once hearing a member of the activist industry say that “peer-reviewed journals are the tool of the enemy” suggesting they were the gold standard communication tool for scientists to report inconvenient facts. At the time I did not appreciate the importance of that statement, and concerningly it appears that this in no longer the case. If we can’t trust the peer-review process to ensure the integrity of papers published in scientific journals, what can we trust? This is a problem that should worry the entire scientific community, not only those concerned with the topic of this particular paper.
